新年快樂!
Happy belated (Lunar) New Year from the Liau Lab! It’s been a really busy past year for us, filled with both science and, well…not-science.
We are all Michelle Yeoh <3
From apple picking to ice skating, we’ve had lots of fun over the past year:
Apple picking, or as the kids like to say, Pick Apple-ing.
Breakfast for dinner at our favorite local IHOP.
Ice skating at the Fenway rink. Brian is a very “down-to-earth” PI.
Enjoying some soondubu at Kaju!
Olivia educates the group on grass jelly desserts at MeetFresh.
Fun aside, we’ve also been hard at work. In fact, 2022-2023 has really been the year of the Liau Lab—well, at least publishing-wise ;) In case you missed our recent scientific work, here’s a quick summary of our papers from the past year. Note: this is continuing off of our previous blog post—go check that out for our recent publications on molecular glue degraders and more!
Lamina-Inducible Methylation and Hi-C (LIMe-Hi-C)
Chemical biology meets 3D genomics! We developed a new approach to simultaneously look at chromosome conformation, DNA methylation, and lamina positioning. Through this, we found and subsequently characterized intriguing Polycomb-marked subcompartments!
DNMT3A base editor scanning
The lab’s first base editor screen, using an innovative reporter approach to study a nonessential gene! We leveraged an endogenous methylation activity reporter to map sequence-activity relationships across DNMT3A. Following up our screen results, we found a noncanonical DNA binding role for the PWWP histone reader domain. What’s more is that our screening strategy can be applied toward other chromatin regulators!
CRISPR-suppressor scanning
Our lab’s official protocol for CRISPR-suppressor scanning is out in Current Protocols! If you’re interested in mapping drug-protein interactions and identifying resistance mutations, you’ll be interested in checking this out.
DNMT1 activity-based CRISPR scanning
CRISPR screens for allostery? Taking advantage of decitabine, an activity-based DNA methyltransferase inhibitor that is nearly identical to the cytosine substrate, we applied CRISPR-suppressor scanning to look for mutations impacting allostery. We found some interesting ones causing DNMT1 hyperactivation. This paper also has some updated and improved analytical tools for analyzing tiling CRISPR screening data.
This paper is currently accepted in principle and we don’t have the pdf yet, but please enjoy our graphical abstract :3
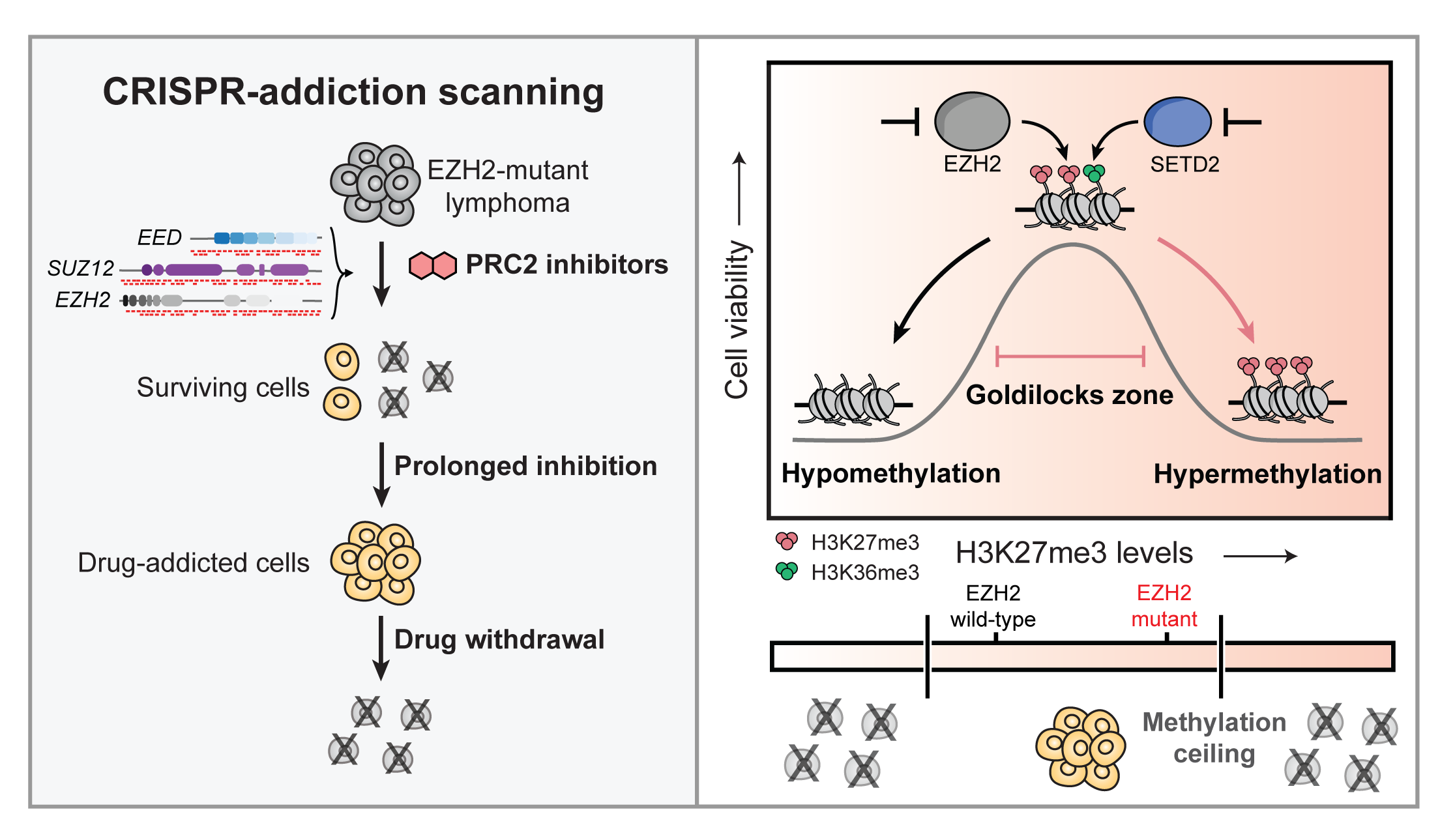
PRC2 drug-addiction scanning
CRISPR screening a whole complex! We used CRISPR-suppressor scanning to systematically mutate the three core members of the PRC2 complex, discovering mutations that confer drug addiction. These pointed toward a model where lymphoma cells need to be in a “Goldilocks” state—too much or too little H3K27me3 is bad.
We had some fun creating art related to some of these studies, though unfortunately they weren’t selected by the journals for the cover/feature. Well, even if journals don’t appreciate our art, hopefully our blog readers will!
Nick, Allison, and Shelby collaborated to make this artistic representation of our LIMe-HiC method. The lime cross-section represents the nucleus, and the red represents GpC methylation.
Nick made this design representing DNMT3A base editor scanning. The pins represent loss-of-function mutations scattered across DNMT3A, and the compass is an homage to base editing.
This past year also saw some new faces arriving in our group. We welcomed the skilled organic chemist, Dr. Stefan Harry, as a joint postdoc with the Bar-Peled lab at MGH. Additionally, we said hi to Tobias Hansen, a visiting graduate student from the University of Copenhagen!
Dr. Stefan Harry
Tobias Hansen
We also welcomed three (count ‘em, yep, we’re popular) new CCB graduate students to the lab:
Calvin Hu
Marc Anthony Zepeda
Idris Barakat
In more bittersweet news, we said goodbye to our longtime lab administrator Rebecca. We’re sad to see her go, but luckily she still works in the department so we can say hi :) On the bright side, we welcomed Lizzy Swenson as our new lab administrator, and have had a great time getting to know her!
Rebecca Stillo
Lizzy Swenson
Continuing with bittersweet departures, our lab has experienced some turnover in our researchers as well. Earlier in the year, we had three talented scientists move on to their next positions:
Pallavi, our first biologist postdoc and a stellar scientist, is now a Senior Scientist at Merck. Pallavi launched the protein biochemistry side of the lab and has provided invaluable expertise to numerous projects. More recently, she led our lab’s study applying CRISPR-suppressor scanning to investigate molecular glues.
Jiaming, a superb synthetic organic chemist postdoc, recently returned to California as a Senior Scientist at Ideaya Biosciences. He served as a mentor and wellspring of expertise for many during his time here, and synthesized a huge number of vital compounds, leaving a lasting mark on our lab (and our freezers).
Sam, a talented research assistant, is now attending medical school at the Cleveland Clinic, Case Western Reserve University! Sam was instrumental in biochemically characterizing DNMT1 mutants and also contributed to numerous other projects.
Check out these videos from Jiaming and Sam’s farewell party, by the way:
You may have seen our last blog post, but we also had four talented graduate students defend (Ally, Kevin, Allison, and Amanda). (*suppresses sobbing*)
Kevin Ngan, Ph.D. Kevin is now working at Tessera Therapeutics.
Allison Siegenfeld, Ph.D. Allison is now doing a postdoc in Prof. Karen Adelman’s lab at HMS.
Amanda Waterbury, Ph.D. Amanda is continuing in our lab as a postdoc for the near future.
Ally Freedy, Ph.D. Ally is now finishing the M.D. part of her M.D.-Ph.D. training at HMS.
With so much going on, 2023 has arrived sooner than any of us expected. But you know what? We have even more cooking, science-wise. Stay on the lookout for the YEAR OF THE LIAU LAB, PART 2!