* indicates equally contributing authors
** indicates corresponding authors
publications and preprints
KBTBD4 cancer hotspot mutations drive neomorphic degradation of HDAC1/2 corepressor complexes. Xie, Xiaowen*; Zhang, Olivia*; Yeo, Megan J.R.*; Lee, Ceejay*; Harry, Stefan A.; Paul, Leena; Li, Yiran; Payne, Connor N.; Nam, Eunju; Kwok, Hui Si; Jiang, Hanjie; Mao, Hiabin; Hadley, Jennifer L.; Lin, Hong; Batts, Melissa; Gosavi, Pallavi M.; D’Angiolella, Vincenzo; Cole, Philip A.; Mazitschek, Ralph; Northcott, Paul A.; Zheng, Ning**; Liau, Brian B.** Submitted.
Asymmetric engagement of dimeric CRL3 KBTBD4 by the molecular glue UM171 licenses degradation of HDAC1/2 complexes. Yeo, Megan J.R.*; Zhang, Olivia*; Xie, Xiaowen*; Nam, Eunju*; Payne, Connor N.; Gosavi, Pallavi M.; Kwok, Hui Si; Iram, Irtiza; Lee, Ceejay; Li, Jiaming; Chen, Nicholas; Jiang, Hanjie; Wang, Zhipeng A.; Lee, Kwangwoon; Mao, Haibin; Harry, Stefan A.; Barakat, Idris A.; Takahashi, Mariko; Waterbury, Amanda L.; Barone, Marco; Mattevi, Andrea; Bar-Peled, Liron; Mazitschek, Ralph; Cole, Philip A.; Liau, Brian B.**; Zheng, Ning** Submitted.
Covalent drug-adduct Grob fragmentation underlies selective LSD1 enzyme inhibition by T-448. Waterbury, Amanda L.*; Caroli, Jonatan*; Zhang, Olivia.; Tuttle, Paloma; Li, Jiaming; Hoenig, Samuel M.; Barone, Marco; Mattevi, Andrea**; Liau, Brian B.** In Revision.
An autoinhibitory switch of the LSD1 disordered region controls enhancer silencing. Waterbury, Amanda L.*; Kwok, Hui Si*; Lee, Ceejay*; Narducci, Domenic N.; Freedy, Allyson M.; Su, Cindy; Raval, Shaunak; Reiter, Andrew H.; Hawkins, William; Lee, Kwangwoon; Li, Jiaming; Hoenig, Samuel M.; Vinyard, Michael E.; Cole, Philip A.; Hansen, Anders S.; Carr, Steven A.; Papanastasiou, Malvina; Liau, Brian B.** Molecular Cell Accepted in Principle.
CTCF/cohesin organize the ground state of chromatin-nuclear speckle association. Yu, Ruofan; Roseman, Shelby A.; Siegenfeld, Allison P.; Nguyen, Son C.; Joyce, Eric F.; Liau, Brian B.; Krantz, Ian D.; Alexander, Katherine; Berger, Shelley L.** bioRxiv 2023. (preprint)
DNA methylation insulates exons from CTCF loops near nuclear speckles. Roseman, Shelby A.*; Siegenfeld, Allison P.*; Lee, Ceejay; Lue, Nicholas Z.; Liau, Brian B.** bioRxiv 2023. (preprint)
DrugMap: A quantitative pan-cancer analysis of cysteine ligandability. Takahashi, Mariko*; Chong, Harrison B*.; Zhang, Siwen*; Lazarov, Matthew J.; Harry, Stefan; Maynard, Michelle; White, Ryan; Murrey, Heather E.; Hilbert, Brendan; Neil, Jason R.; Gohar, Magdy; Ge, Maolin; Zhang, Junbing; Durr, Benedikt R.; Kryukov, Gregory; Tsou, Chih-Chiang; Brooijmans, Natasja; Alghali, Aliyu; Rubio, Karla; Vilanueva, Antonio; Harrison, Drew; Koglin, Ann-Sophie; Ojeda, Samuel; Karakyriakou, Barbara; Healy, Alexander; Assaad, Jonathan; Makram, Farah; Rachman, Inbal; Khandelwal, Neha; Tien, Pei-Chieh; Popoola, George; Chen, Nicholas; Vordermark, Kira; Richter, Marianne; Patel, Himani; Yang, Tzu-Yi; Griesshaber, Hanna; Hosp, Tobias; van den Ouweland, Sanne; Hara, Toshiro; Bussema, Lily; Dong, Rui; Shi, Lei; Rasmussen, Martin Q.; Domingues, Ana C.; Lawless, Aleigha; Fang, Jacy; Yoda, Satoshi; Nguyen, Linh P.; Reeves, Sarah M.; Wakefield, Farrah N.; Acker, Adam; Clark, Sarah E.; Dubash, Taronish; Fisher, David E.; Maheswaran, Shyamala; Haber, Daniel A.; Boland, Genevieve; Sade-Feldman, Moshe; Jenkins, Russel; Hata, Aaron; Bardeesy, Nabeel; Suva, Mario L.; Martin, Brent; Liau, Brian B.; Ott, Christopher; Rivera, Miguel N.; Lawrence, Michael S.**; Bar-Peled, Liron**. Cell 2024, 187, 2536–2556. (link)
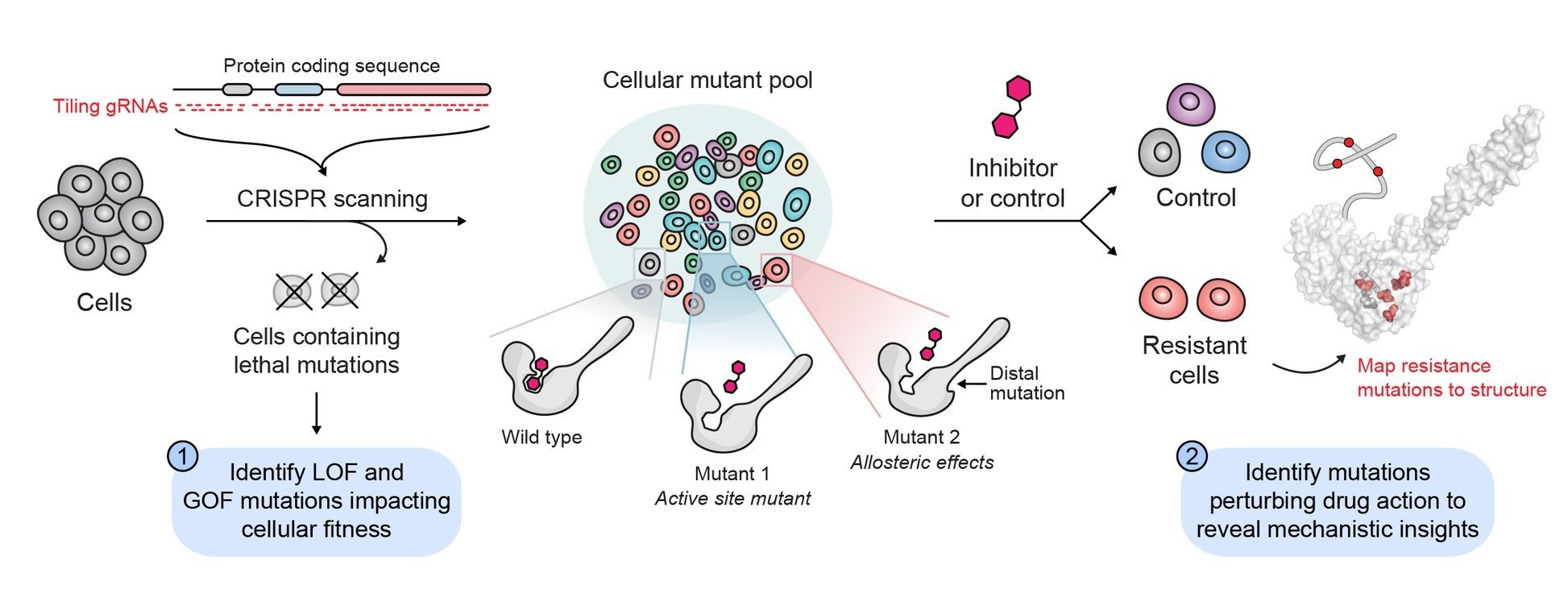
Base editor screens for in situ mutational scanning at scale.
Lue, Nicholas Z.; Liau, Brian B.** Molecular Cell 2023, 83, 2167–2187. (link) Review
Coverage: Featured on the Cover of Molecular Cell
Systematic identification of anticancer drug targets reveals a nucleus-to-mitochondria ROS-sensing pathway.
Zhang, Junbing**; Simpson, Claire M.; Berner, Jacqueline; Chong, Harrison B.; Fang, Jiafeng; Ordulu, Zehra; Weiss-Sadan, Tommy; Possemato, Anthony P.; Harry, Stefan; Takahashi, Mariko; Yang, Tzu-yi; Richter, Marianne; Patel, Himani; Smith, Abby E.; Carlin, Alexander D.; Hubertus de Groot, Adriaan F.; Wolf, Konstantin; Shi, Lei; Wei, Ting-Yu; Dürr, Benedikt R.; Chen, Nicholas J.; Vornbäumen, Tristan; Wichmann, Nina O.; Mahamdeh, Mohammed S.; Pooladanda, Venkatesh; Matoba, Yusuke; Kumar, Shaan; Kim, Eugene; Bouberhan, Sara; Oliva, Esther; Rueda, Bo R.; Soberman, Roy J.; Bardeesy, Nabeel; Liau, Brian B.; Lawrence, Michael; Stokes, Matt P.; Beausoleil, Sean A.; Bar-Peled, Liron**. Cell 2023, 186, 2361–2379. (link)
Drug addiction unveils a repressive methylation ceiling in EZH2-mutant lymphoma.
Kwok, Hui Si*; Freedy, Allyson M.*; Siegenfeld, Allison P.; Morriss, Julia W.; Waterbury, Amanda L.; Kissler, Stephen M.; Liau, Brian B.** Nature Chemical Biology 2023, 19, 1105–1115. (link) (preprint)
Resources: GitHub
Coverage: Nature Chemical Biology News & Views, Trends in Pharmacological Sciences Spotlight
Activity-based CRISPR scanning uncovers allostery in DNA methylation maintenance machinery.
Ngan, Kevin C.; Hoenig, Samuel M.; Kwok, Hui Si; Lue, Nicholas Z.; Gosavi, Pallavi M.; Tanner, David A.; Garcia, Emma M.; Lee, Ceejay; Liau, Brian B.** eLife 2023, 12, e80640. (link) (preprint)
Resources: GitHub
Chromatin complex dependencies reveal targeting opportunities in leukemia.
Najm, Fadi J.; DeWeirdt, Peter; Moore, Molly M.; Bevill, Samantha M.; El Farran, Chadi A.; Macias, Kevin A.; Hegde, Mudra; Waterbury, Amanda L.; Liau, Brian B.; van Galen, Peter; Doench, John G.; Bernstein, Bradley E.** Nature Communications 2023, 14, 448. (link)
Profiling the landscape of drug resistance mutations in neosubstrates to molecular glue degraders.
Gosavi, Pallavi M.*; Ngan, Kevin C.*; Yeo, Megan J.R.; Su, Cindy; Li, Jiaming; Lue, Nicholas Z.; Hoenig, Samuel M.; Liau, Brian B.** ACS Central Science 2022, 8, 417–429. (link) (preprint)
Coverage: ACS Central Science First Reactions
Discovering new biology with drug-resistance alleles.
Freedy, Allyson M.; Liau, Brian B.** Nature Chemical Biology 2021, 17, 1219–1229. (link) Review
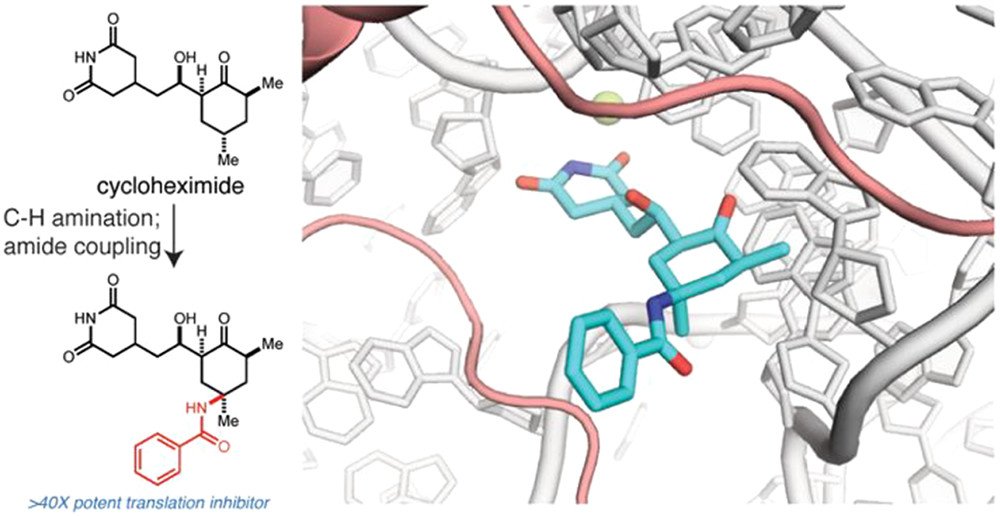
Discovery of C13-aminobenzoyl cycloheximide derivatives that potently inhibit translation elongation.
Koga, Yumi*; Hoang, Eileen M.*; Park, Yongho; Keszei, Alexander F.A.; Murray, Jason; Shao, Sichen**; Liau, Brian B.** Journal of the American Chemical Society 2021, 143, 13473–13477. (link)
Coverage: Synfacts
Taking a STAP at core promoter–transcriptional cofactor specificity.
Siegenfeld, Allison P.*; Lee, Ceejay*; Liau, Brian B.** Biochemistry 2019, 58, 3133-3135. (link) Invited Commentary
CRISPR-suppressor scanning reveals a nonenzymatic role of LSD1 in AML.
Vinyard, Michael E.*; Su, Cindy*; Siegenfeld, Allison P.*; Waterbury, Amanda L.*; Freedy, Allyson M.*; Gosavi, Pallavi M.*; Park, Yongho; Kwan, Eugene E.; Senzer, Benjamin D.; Doench, John G.; Bauer, Daniel E.; Pinello, Luca; Liau, Brian B.** Nature Chemical Biology 2019, 15, 529–539. (link)
A versatile synthetic route to cycloheximide and analogues that potently inhibit translation elongation.
Park, Yongho; Koga, Yumi; Su, Cindy; Waterbury, Amanda L.; Johnny, Christopher L.; Liau, Brian B.** Angewandte Chemie International Edition 2019, 58, 5387–5391. (link)
Uncovering the cellular target of agelastatin A.
Park, Yongho; Liau, Brian B.** Cell Chemical Biology 2017, 24, 542–543. (link) Invited Commentary
prior to liau lab
Gain-of-function genetic alterations of G9a drive oncogenesis. Kato, S., Weng, Q.Y., Insco, M.L., Chen, K.Y., Muralidhar, S., Pozniak, J., Diaz, J.M.S., Drier, Y., Nguyen, N., Lo, J.A., van Rooijen, E., Kemeny, L.V., Zhan, Y., Feng, Y., Silkworth, W., Powell, C.T., Liau, B.B., Xiong, Y., Jin, J., Newton-Bishop, J., Zon, L.I., Bernstein, B.E., Fisher, D.E. Cancer Discovery 2020, 10, 980–997.
Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Miller, T.E., Liau, B.B., Wallace, L.C., Morton, A.R., Xie, Q., Dixit, D., Factor, D.C., Kim, L.Y.J., Morrow, J.J., Wu, Q., Mack, S.C., Hubert, C.G., Gillespie, S.M., Flavahan, W.A., Hoffmann, T., Thummalapalli, R., Hemann, M.T., Paddison, P.J., Horbinski, C.M., Zuber, J., Scacheri, P.C., Bernstein, B.E., Tesar, P.J., Rich, J.N. Nature 2017, 547, 355-359.
Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Liau, B.B.*, Sievers, C.*, Donohue, L.K., Gillespie, S.M., Flavahan, W.A., Miller, T.E., Venteicher, A.S., Hebert, C.H., Carey, C.D., Rodig, S.J., Shareef, S.J., Najm, F.J., van Galen, P., Wakimoto, H., Cahill, D.P., Rich, J.N., Aster, J.C., Suvà, M.L., Patel, A.P., Bernstein, B.E. Cell Stem Cell 2017, 20, 233–246.
RBPJ maintains brain tumor-initiating cells through CDK9-mediated transcriptional elongation. Xie, Q., Wu, Q., Kim., L., Miller, T.E., Liau, B.B., Mack, S.C., Yang, K., Factor, D.C., Fang, X., Huang, Z., Zhou, W., Alazem, K., Wang, X., Bernstein, B.E., Bao, S., Rich, J.N. Journal of Clinical Investigation 2016, 126, 2757–2772.
Insulator dysfunction and oncogene activation in IDH mutant gliomas. Flavahan, W.A.*, Drier, Y.*, Liau, B.B., Gillespie, S.M., Venteicher, A.S., Stemmer-Rachamimov, A.O., Suvà, M.L., Bernstein, B.E. Nature 2016, 529, 110–114.
A multiplexed system for quantitative comparisons of chromatin landscapes. van Galen, P., Viny, A.D., Ram, O., Ryan, R.J.H., Cotton, M.J., Donohue, L., Sievers, C., Drier, Y., Liau, B.B., Gillespie, S.M., Carroll, K.M., Cross, M.B., Levine, R.L., Bernstein, B.E. Molecular Cell 2016, 61, 170–180.
Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Pelish, H.E.*, Liau, B.B.*, Nitulescu, I.I., Tangpeerachaikul, A., Poss, Z.C., Da Silva, D.H., Caruso, B.T., Arefolov, A., Fadeyi, O., Christie, A.L., Du, K., Banka, D., Schneider, E.V., Jestel, A., Zou, G., Si, C., Ebmeier, C.C., Bronson, R.T., Krivtsov, A.V., Myers, A.G., Kohl, N.E., Kung, A.L., Armstrong, S.A., Lemieux, M.E., Taatjes, D.J., Shair, M.D. Nature 2015, 526, 273–276.
A unified strategy for the synthesis of 7-membered-ring-containing Lycopodium alkaloids. Lee, A.S., Liau, B.B., Shair, M.D. Journal of the American Chemical Society 2014, 136, 13442–13452.
Total syntheses of HMP-Y1, hibarimicinone, and HMP-P1. Liau, B.B., Milgram, B.C., Shair, M.D. Journal of the American Chemical Society 2012, 134, 16765–16772.
Gram-scale synthesis of the A'B'-subunit of angelmicin B. Milgram, B.C., Liau, B.B., Shair, M.D. Organic Letters 2011, 13, 6436–6439.
Total synthesis of (+)-fastigiatine. Liau, B.B., Shair, M.D. Journal of the American Chemical Society 2010, 132, 9594–9595.
A study of the effect of Z- and E-olefinic geometry on the rates of ring closure of 13-membered dienes. Liau, B.B., Gnanadesikan, V., Corey, E.J. Organic Letters 2008, 10, 1055–1057.